The profitable poultry production is often influenced by various factors among which infectious diseases remains as a constant and critical challenge. The emergence and reemergence of novel pathogens are potential threat for the effective management of diseases in poultry farms and consequently the productivity. The turnaround time (TAT) is defined as the time interval between the specimens received in the laboratory to the time of reports dispatched with verification. The TAT for the traditional conventional diagnostic assays involving isolation of the pathogen, serological assays etc., generally range from two to seven days() although the sensitivity and specificity could be analogous with molecular diagnostics. The advent of molecular diagnostics targeting the genomic material unique to each pathogen have become the game changer underscored by the time-sensitive nature of many of the treatment decisions associated with infectious diseases (Scott Powell, 2015). Hence, the availability of a rapid, accurate, specific and sensitive diagnostic assay is invaluable during an epidemic for the veterinarians to decide on the appropriate prevention and control strategies to obviate the economic loss to the farmers.

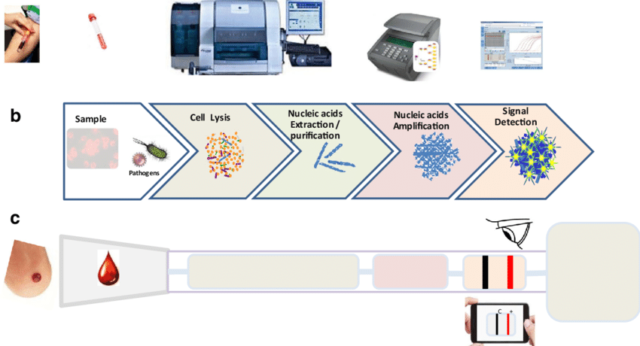
Molecular Diagnosis/ Nucleic Acid-Based Amplification Techniques
The progress in the culture-independent assays like genomics and proteomics based approaches facilitate to discriminate the closely related species as well as to determine the phylogenetic relationship of the isolates. The sensitivity and specificity for all the molecular tests are governed by three critical control points, including nucleic acid extraction, enzymes used for amplification and the sequence of primers and probes (David. et al2007)
Advantages:
- Rapid, Sensitive and specific
- Cheapand simultaneous detection of multiple etiology
- Detection of pathogens that are uncultivable or difficult and harmful to culture, slow growing viruses in culture, pathogens displaying antigenic variations
- No hazard of handling live pathogen and its dissemination
- Shipping of the sample in an inactivated form (FTA Cards)
- Detects the pathogens in formaldehyde-fixed paraffin-embedded (FFPE) material, body fluids, eggs, feathers, swabs and environmental samples such as dust, soil or litter
Polymerase Chain Reaction (PCR)
PCR is a characteristic example of nucleic acid amplification assay that has revolutionized the field of molecular diagnosis since developed by Mullis and Faloona. PCR is based on extraction and purification of DNA molecule and exponential amplification of the target sequence, using a thermostable DNA polymerase and two specific oligonucleotide primers. After the PCR reaction, the amplified product can be detected by several techniques, including gel electrophoresis, colorimetric methods, and sequencing. PCR has been used for the detection pathogens with overall sensitivity and specificity ranging from 77.8% to 100% and 89% to 100% respectively. Hence, PCR can be employed for the detection of pathogens in a variety of specimen types. In addition to the conventional PCR, more advanced variants of the technique are in vogue.
Reverse Transcription-PCR (RT-PCR)
RT-PCR was designed to amplify RNA targets. In this technique, reverse transcriptase (RT) is used to convert viral RNA targets into complementary DNA (cDNA), and then the resulting cDNA is amplified by conventional PCR for the detection of infection caused by RNA viruses. RT-PCR demonstrated overall sensitivity and specificity ranging from 73% to 100% and 99% to 100% respectively in the detection of viral infection.
Real-Time PCR
In real-time PCR system, viral nucleic acid amplification and detection steps are carried out at the same time. The detection of the amplification product is relied on the amount of fluorescence emission from the specimen. The fluorescence emission from the specimen is monitored by special thermal cycler. The computer, with appropriate software connected to the thermal cycler, records the data and produces an amplification plot at every reaction cycle. The detection and quantification of amplification products can be done by using SYBR green, the Taq Man, and molecular beacon chemistries.
The SYBR green dye binds to the minor groove of double-stranded DNA (dsDNA) product and upon excitation by appropriate light, it exhibits improved fluorescence, which is directly proportional to the accumulated dsDNA product.
The Taq Man probe is a DNA oligonucleotide with a fluorescent dye termed reporter attached to fluorophore one end (5′ base) and quencher on the other (3′ base). Taq Man probes are designed to hybridize to an internal region of a PCR product. During the annealing stage of the PCR, both the primer and the Taq Man probe bind to the template strand. When the Taq DNA polymerase extends the primer, the polymerase cleaves the probe by its the 5′-3′ exonuclease activity. Cleavage of the probe leads to the release of the fluorescent dye resulting in fluorescence emission. The amount of fluorescence is directly proportional to the PCR product.
Molecular beacon is a small DNA molecule with a fluorescent dye at the 5′ end and a quencher at the 3′ end. The sequences at the very 3′ and 5′ ends are complementary to each other. The internal part of the molecule is designed to be complementary to the target sequence of a PCR product. When molecular beacon is free in solution, it will adopt a hairpin structure. This brings the fluorophore and quencher in close proximity, leading to absorption of emitted light of the florescent dye by the quencher and hence fluorescence is not detected. However, when a molecular beacon hybridizes to the target sequence, the fluorophore and quencher are separated, leading to the emission of fluorescence. The amount of fluorescence is directly proportional to the PCR product.
Transcription-Based Amplification Methods
Transcription-based amplification methods have several advantages, since they do not require a thermal cycler, so developing countries and budget-restricted laboratories can afford to perform the assays, they have rapid kinetics (requires fewer cycles), and they produce a single-stranded RNA product that is suitable for detection by various techniques. Transcription-based amplification methods are suitable for the diagnosis of viral infections caused by RNA viruses. They can amplify viral genomic RNA, messenger RNA, or ribosomal RNA.
This includes nucleic acid sequence-based amplification (NASBA) and transcription-mediated amplification (TMA). NASBA and TMA are similar to each other. They are isothermal amplification methods. The entire amplification process is carried out at the temperature of 41°C. In both cases, the viral RNA target is first converted into cDNA with RT and then RNA polymerase synthesizes multiple copies of viral RNA product. The only difference between TMA and NASBA in the amplification process is two enzymes (RT and RNA polymerase) are utilized in case of TMA while NASBA utilizes three enzymes (avian my eloblastosis virus reverse transcriptase (AMV-RT), RNase H, and T7 RNA polymerase).
In NASBA, three enzymes and two primers work together to exponentially amplify a target viral RNA. Primer 1 (P1) carries at its 5′ end T7 RNA polymerase promotor region and at its 3′ end, P1 carries sequence that is complementary to a target viral RNA sequence. Primer 2 (P2) carries a sequence complementary to cDNA strand. The amplification reaction begins with the production of cDNA copy of the viral RNA by RT using P1. RNase H degrades the viral RNA from RNA-DNA hybrid molecules. Then, RT synthesizes dsDNA molecules using P2 and the released DNA strand. Finally, T7 RNA polymerase uses dsDNA molecules as templates to transcribe many viral RNA copies. The cycle is repeated several times, resulting in the accumulation of many viral RNA copies and ds DNA molecules. The amplified product can either be detected by gel electrophoresis at the end of the assay or in real time using molecular beacon.
Loop-Mediated Isothermal Amplification (LAMP)
LAMP is another isothermal nucleic acid amplification method that is extensively utilized for sensitive, specific, rapid, and cost-effective detection of both DNA and RNA clinical specimens. The method employs four to six unique primers which include the forward inner primer (FIP), backward inner primer (BIP), forward outer primer (F3), and backward outer primer (B3) and DNA polymerase with strand-displacement activity to amplify target DNA. The addition of RT in LAMP reaction (RT-LAMP) permits the amplification of RNA target. The primers are specifically designed to recognize six precise regions from a targeted nucleic acid sequence. In addition to the above primers, two more loop primers, namely, forward loop primer (LF) and backward loop primer (LB) were included to accelerate LAMP assay. Due to the use of four to six specific primers, LAMP assay has outstanding sensitivity and specificity in the detection of target nucleic acids. The LAMP reaction is performed in constant temperature between 60–65°C, without the need for expensive specialized equipment. The method requires only inexpensive heating block or water bath, making it very useful under poor laboratory settings. The LAMP reaction takes turnaround time of less than 1 hour and the amplified product can be detected by several methods, including the real-time measurement of the turbidity caused by precipitated magnesium pyrophosphate using a tur bido meter, visual detection of magnesium pyrophosphate precipitation following completion of the reaction, detection of fluorescence under ultraviolet light or natural light by adding an intercalating fluorescent dye to the final reaction mixture, and visualization of the bands with various sizes using agarose gel electrophoresis.
DNA Microarrays
In DNA microarray diagnosis, fluorescently labeled viral nucleic acids in a test sample are used to screen an array of oligonucleotide probes immobilized on a solid surface (e.g., glass slide). The oligonucleotide probes used here are specific for the genome of the target virus. The results of hybridization between immobilized probes and target sequences labeled with fluorescent dyes are detected and quantified by fluorescence-based detection. It is a high-throughput tool as it allows multiplex detection of a large number of potential viral pathogens in clinical specimens. The limitations of the technique includes, too expensive to be used for routine clinical diagnosis, labor-intensive, and time-consuming (the hybridization process may take hours to days to complete). Nonspecific hybridization between test materials and immobilized probes can affect the sensitivity of the assay. In addition, designing of specific probes requires almost complete information of the genetic makeup the virus of interest. The assay detects only those viral pathogens that have target probes on the array.
Next-Generation Sequencing (NGS)
NGS involves preparation of test sample, sequencing of the target nucleic acid fragments using one of the available NGS platforms, and analysis of the sequence data using suitable bioinformatic tools. Several companies produce different NGS machines that use different methods of sequencing, reagents, and data analysis tools. Eg. , Pyrosequencing (Roche 454) detects release of pyrophosphate following incorporation of nucleotides in a DNA polymerization process. Illumina’s NGS platforms detect release of fluorescent labels from incorporated nucleotides in a DNA polymerization process. The emerging technologies like Oxford nanopore (MinIon) platform sequences the target nucleic acid by sensing the ionic current of DNA/RNA molecules that pass through the nanopores. Unlike PCR and DNA microarray methods, NGS does not require prior knowledge of genomic sequences of the viral pathogens. It does not also require target specific PCR primers and oligonucleotide probes. But the limitations like the turnaround time, the number of samples per run, cost of sequencers, and requirement of skills in bioinformatics are the constraints in its wider usage.
Dr. K.Ramya and Dr. K.Sukumar
Department of Veterinary Microbiology,
Veterinary College and Research Institute, Namakkal – 637 002
Tamil Nadu Veterinary and Animal Sciences University
E mail: ramyasankar83@gmail.com
















