
Bacteria were first observed and described by Antonie van Leeuwenhoek in 1676 when he published his observations in a series of letters to the Royal Society.
Now, centuries later it is estimated that there are approximately 1 trillion microbial species on earth – a diverse population with major differences between them. If you would compare this number to all plant and animal species combined (approximately 10 million species), then we can conclude that we live in a bacterial world.
Everywhere around us we are surrounded by bacteria. Humans and animals have an interesting relationship with bacteria. For instance, we rely on them as bacteria help us with the digestion of fibres in our intestinal tract – fibres that we cannot digest ourselves.
Mammals rely on microbes to digest these for them instead. There is a clear mutual benefit in this relationship.
On the other hand, we also might fear them as some microbial species can be pathogenic and can cause diseases. It all depends on the species and strains of microbes. Strains matter.
Microbes (like all species) are described in different levels from Domain, Phylum, Class, Order, Family, Genus down to Species. In Table 1 the classification steps are shown for bacteria (left) and for humans (right).
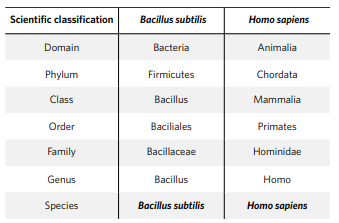
Table 1. Classification steps for bacteria and humans. In the human case it ends with Homo Sapiens at species level. However, the same steps take the bacterial definition down to Bacillus subtilis level, while there is a bigger variety of microbes than people in a species level. Providing microbes with a strain code makes it possible to identify which Bacillus subtilis is being referred to. Microbe strains and their characteristics differ significantly from each other within a species level. Within Bacillus subtilis, there can be thousands of different strains which can be considered as unique individuals. Strains can be considered as individuals within a population. Consequently, a colony of strains is then like a clone population of the same individual.
A strain is a subtype of a microbial species with:
- Unique genetic identity
- Distinctive morphological, biochemical and behavioural features
For instance, the bacterial species Bacillus subtilis consists of more than 1,000 strains that are recorded in international strain banks. In contrast: the 250 cattle, 190 dog, and 65 chicken recognised breeds together are not close to the 1,000 Bacillus subtilis diversity within one species. A few examples are provided to highlight some of the diversity found within Bacillus subtilis.
EXAMPLE 1
Enzyme production characteristics of Bacillus subtilis Cellulase and xylanase are two fibre degrading enzymes needed to break down cellulose and hemicellulose, respectively. Their activity varies exponentially across different Bacillus strains.
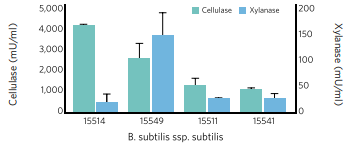
Bacillus strains differ significantly within the Bacillus subtilis group in terms of cellulase and xylanase activity. This shows that not all Bacillus subtilis will therefore have the same effect when fed to an animal as a probiotic.
EXAMPLE 2
High genetic diversity within Bacillus subtili. In the same category or function, genes of the same strain or species can be identified as divergent if their expression would lead to a different outcome.
The number of genes associated with different category groups and the levels of total divergence among a certain strain identify diversity can be evaluated by genotyping. In Fig. 2, 17 different Bacillus subtilis were genotyped (Earl et al. (2017) in order to understand the level of genetic similarity for a certain function.
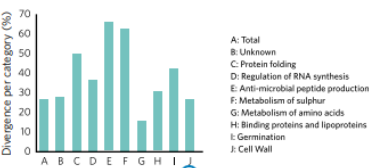
Traits crucial for the survival of the strain were identified such as: cell wall production, metabolism, germination, etc. We can see a very high diversity of genetic divergence for most of the function, from 16% for metabolism of amino-acids to even 66% of divergence for anti-microbial peptide production.
EXAMPLE 3
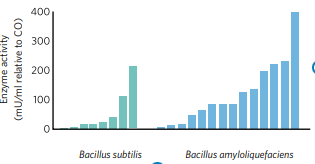
Pathogen inhibition activity To test inhibitory effects against Salmonella heidelberg an in vitro pathogen inhibition test was conducted in Denmark (EXP-18-AH5439).
Salmonella heidelberg was plated in agar and Bacillus strains were inoculated to see their inhibitory effects to this pathogen. Inhibition zones were measured. The inhibitory activity of different Bacillus subtilis strains against the pathogenic bacteria Salmonella heidelberg varies greatly. Some Bacillus strains have no effect on Salmonella inhibition, while others do.
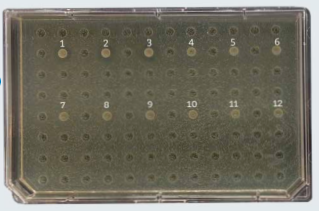
In Fig. 3, we can visualise the pathogen inhibition effect of some Bacillus strains from Chr. Hansen bank of strains vs other Bacillus strains present on the market.
Conclusion
Assessing bacterial genetic diversity is critical for strain identification. Selecting and differentiating strains is the basis for success as different strains all have unique properties. When the right strains are selected for the right target solution they can be an effective solution to counter major food and health challenges.
There is not one strain that is effective against each problem. It is a matter of selecting the right strains or combination of strains for the right objective.
Chr. Hansen recently selected three Bacillus strains in a new product for the poultry market: GALLIPRO® Fit. The foundation was the selection and combination of strains that were most effective for the poultry industry. The strains were selected due to their strong pathogen inhibition and enzyme production abilities.
Strains matter. Selected combinations of strains is key to a successful solution from which poultry farmers can benefit.
By: Christophe Bostvironnois, DVM – Chr. Hansen, Hoersholm, Denmark Roland Koedijk, PhD









